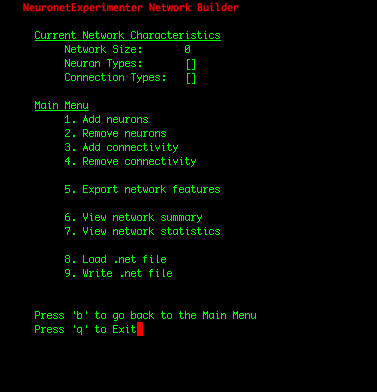

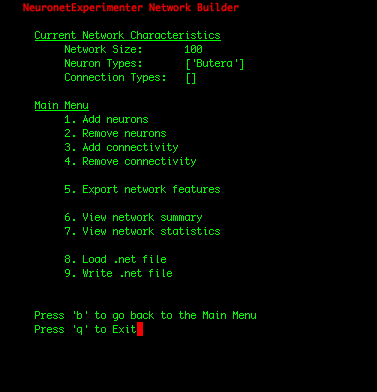
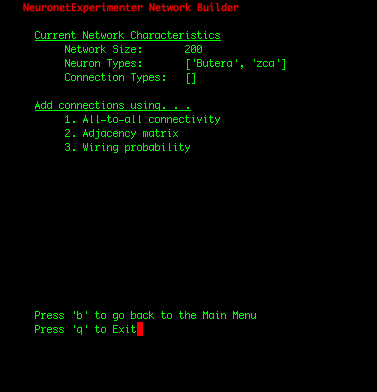
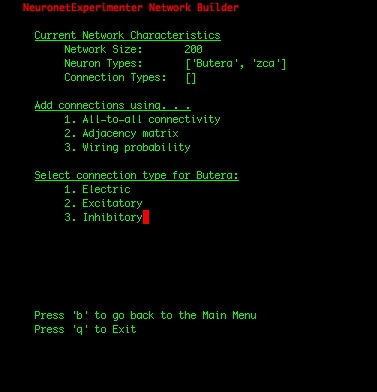
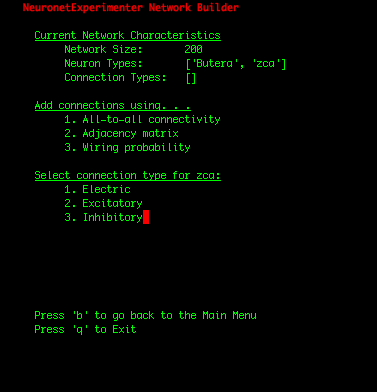
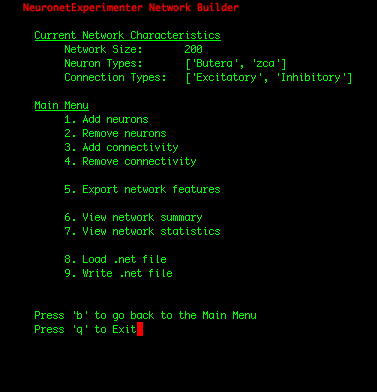
| IMPORTANT: Since the first 100 neurons in our network were Butera neurons and the later 100 neurons were zca neurons (which we can observe by selecting the "View network summary" option in the Main Menu), we had to construct the adjacency matrix in such a manner that assures that the first 100 rows correspond to connections FROM the Butera neurons, and the later 100 rows represent connections FROM the zca neurons. If the rows of the adjacency matrix should be mapped to the neuron types in a more complicated arrangement, one solution is to write a simple shell script that adds individual neurons to your network in the order you need them using the Utility Scripts. Or for Python programmers, the cleanest solution may be to add the neurons using the Network class in the nne.py library that the Utility Scripts themself use. Here is a simple example. |