This sets each neuron's g_syn value to 0 for the first second of the simulation (i.e., 2 iterations of the 500 ms long 'unconnected' experiment) and then switches back to the original g_syn for the rest of the simulation. The results look like this:
| Network
zca.net
Dt 0.05 Print Step Size 1 Output V hhs nns bbs zzs s_AMPA Experiment control 1 Change g_NaP = 0 # Sets the g_NaP value for each neuron to zero Change g_A *= 2.0 # Doubles whatever the current g_A value is for that neuron Change g_syn = gauss(1.0, 0.25) # Changes all the neurons in the network so that g_syn is a value pulled # from a Gaussian distribution with mean of 1.0 and standard # deviation of 0.25. |
| Network
zca.net
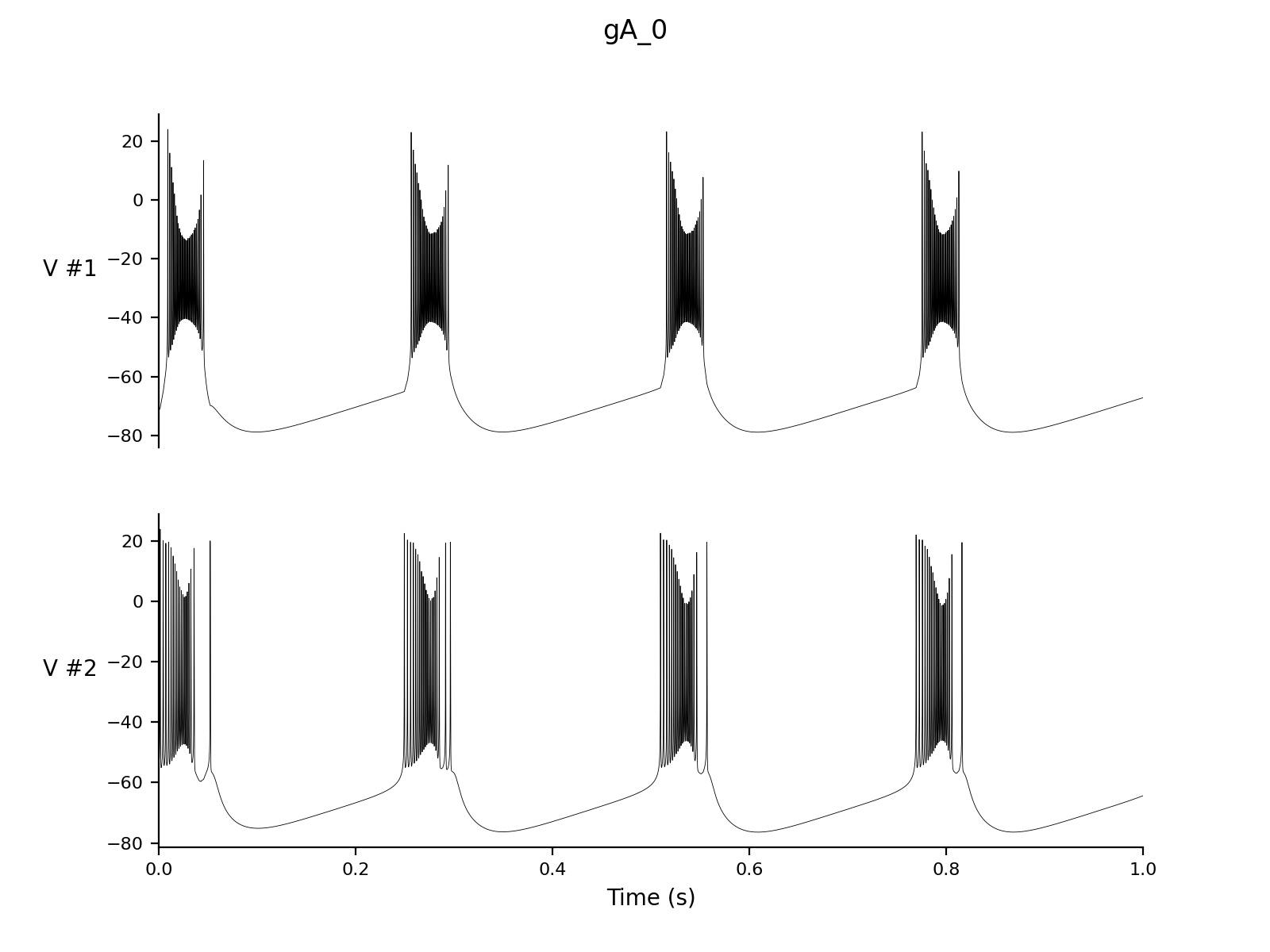
Dt 0.05 Print Step Size 1 Output V hhs nns bbs zzs s_AMPA Experiment control 1 Change g_A = 0 |
| Network
zca.net
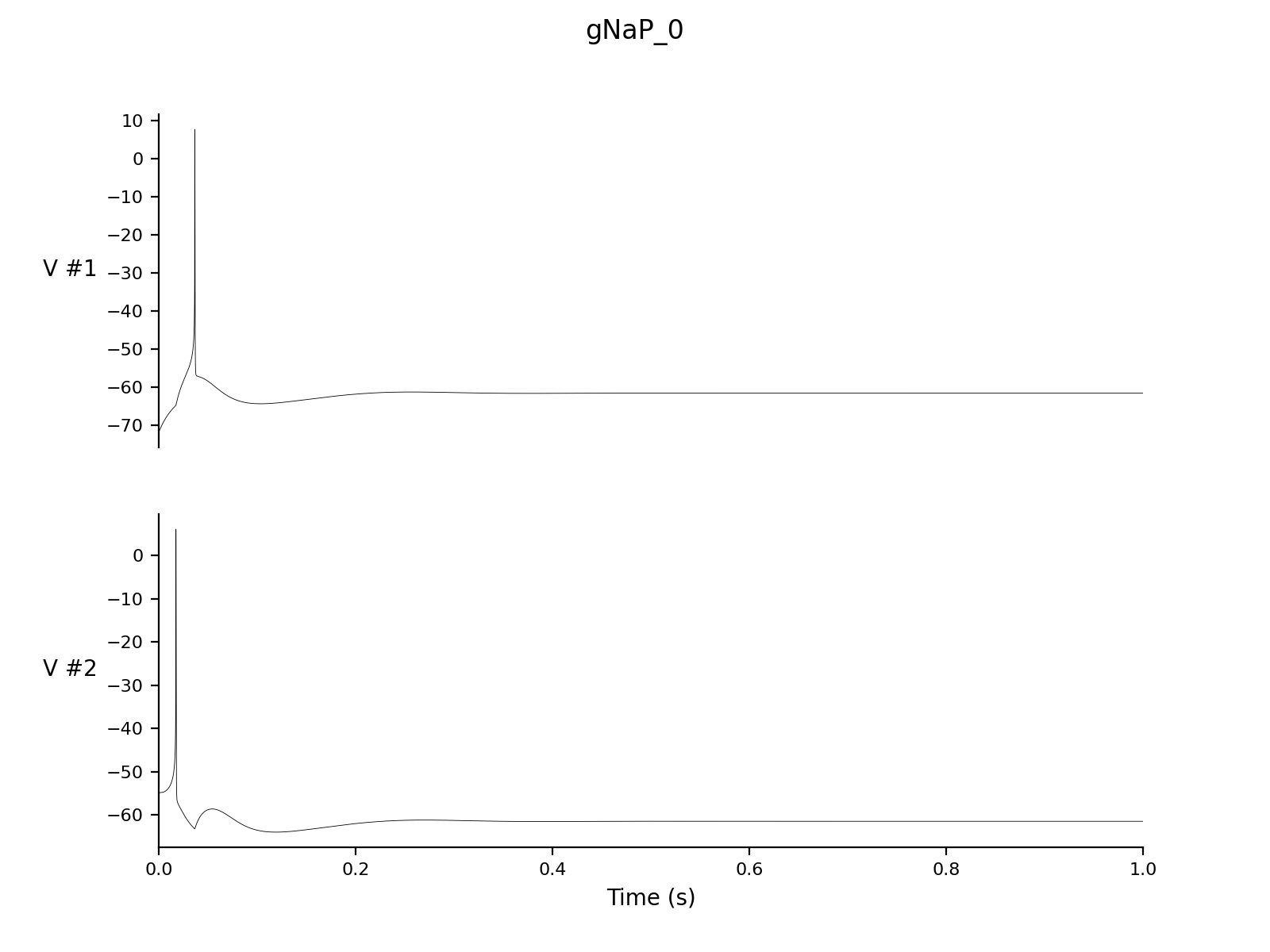
Dt 0.05 Print Step Size 1 Output V hhs nns bbs zzs s_AMPA Experiment control 1 Change g_NaP = 0 |
| Network
zca.net
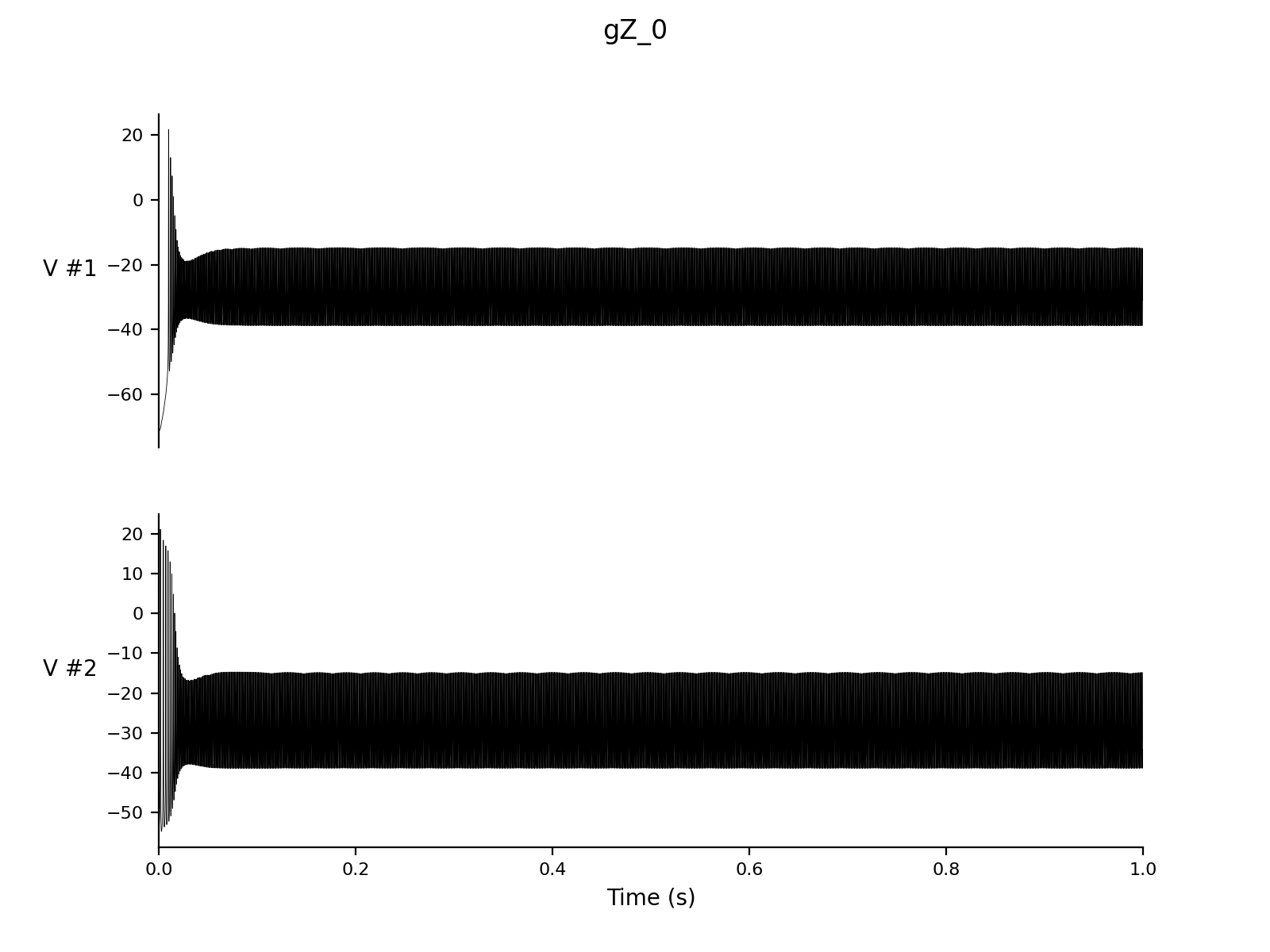
Dt 0.05 Print Step Size 1 Output V hhs nns bbs zzs s_AMPA Experiment control 1 Change g_Z = 0 |
| >
cd simulations/ca1 > nne_run gA_0.setup > nne_run gNaP_0.setup > nne_run gZ_0.setup |
| > nne_plotPair
gA_0.setup 1 2 |
| Network
zca.net
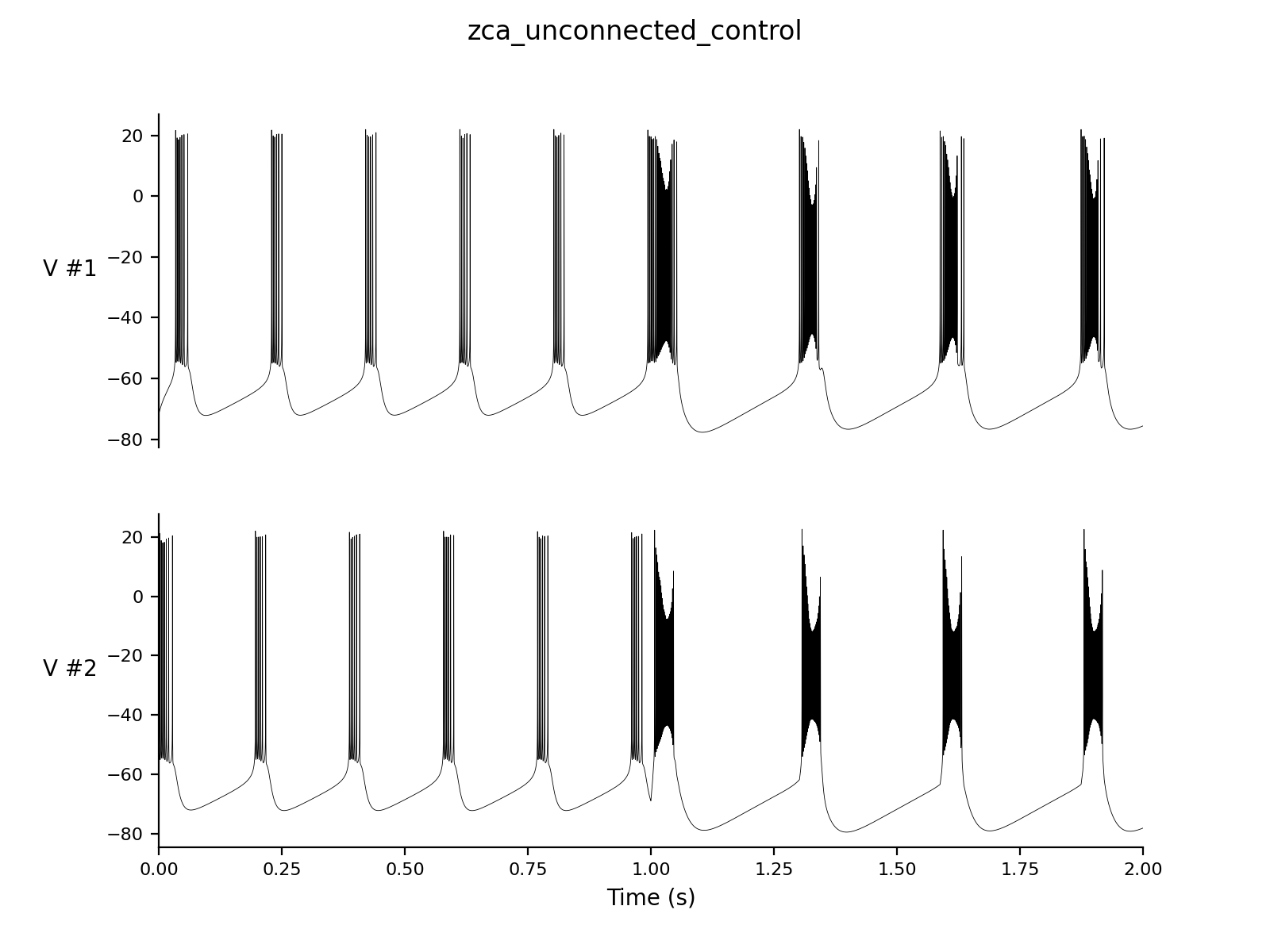
Dt 0.05 Print Step Size 1 Output V hhs nns bbs zzs s_AMPA Experiment unconnected 2 control 2 |
| Time:
500
Changes g_syn = 0 |
| Time:
500
|
