| #
string user uses to refer to this connection type :
list of variables that are connected by this connection
type, e.g., # Excitatory: s_AMPA s_NMDA # Inhibitory: s_GLY s_GABA # Electrical: V Excitatory: s_AMPA |
| #
Excitatory cortical neurons, 0 [Ca]. # number C_m=1.0 number pms=3,pns=4 number V_Na=55.0,tau_h=-40.5,tau_n=-27.0 number theta_a=-50.0,sigma_a=20.0,theta_b=-80.0,sigma_b=-6.0,tau_Bs=15.0 number sigma_m=9.5,sigma_h=-7.0,sigma_n=10.0,sigma_z=5.0,sigma_k=7 # p g_Na=35.0,g_Kdr=6.0,g_L=0.05,I_app=0.661914 p g_A=1.4,g_NaP=0.3,g_Z=1.0 p theta_z=-39.0,tau_Zs=75.0 p phi=10.0, theta_h=-45.0 p theta_m=-30.0,theta_n=-35.0,theta_p=-47.0,sigma_p=3.0 p V_K=-90.0,V_L=-70.0 p g_syn=0, tau_s=15, theta_s=-10, sigma_s=-2 # GAMMAF(VV,theta,sigma)=1.0/(1.0+exp(-(VV-theta)/sigma)) # V'=(-g_L*(V-V_L)-I_Na-I_NaP-I_Kdr-I_A-I_z-I_syn+I_appx)/C_m hhs'=phi*(GAMMAF(V,theta_h,sigma_h)-hhs)/(1.0+7.5*GAMMAF(V,tau_h,-6.0)) nns'=phi*(GAMMAF(V,theta_n,sigma_n)-nns)/(1.0+5.0*GAMMAF(V,tau_n,-15.0)) bbs'=(GAMMAF(V,theta_b,sigma_b)-bbs)/tau_Bs zzs'=(GAMMAF(V,theta_z,sigma_z)-zzs)/tau_Zs s_AMPA'=((1-s_AMPA)*s_inf(V)-s_AMPA)/tau_s # I_appx=I_app #Iappx=if(t<=3.0)then(Iapp)else(0.0) Minfs=GAMMAF(V,theta_m,sigma_m) Pinfs=GAMMAF(V,theta_p,sigma_p) Ainfs=GAMMAF(V,theta_a,sigma_a) s_inf(V)=1./(1.+exp((V-theta_s)/sigma_s)) # I_Na=g_Na*(Minfs^pms)*hhs*(V-V_Na) I_NaP=g_NaP*Pinfs*(V-V_Na) I_Kdr=g_Kdr*(nns^pns)*(V-V_K) I_A=g_A*Ainfs^3*bbs*(V-V_K) I_z=g_Z*zzs*(V-V_K) I_syn=g_syn*sum(s_AMPA)*V # init V=-71.81327 init hhs=0.98786 init nns=0.02457 init bbs=0.203517 init zzs=0.00141 init s_AMPA=0.0 |
| > nne_build |
| Network
zca.net
# Indicates the simulator should use the network in
zca.net Dt 0.05 # with a timestep of 0.05 Print Step Size 1 # and should output data every time step Output V hhs nns bbs zzs s_AMPA # and should output time and each of the state variables for all the neurons Method: rk4 Setup Seed 1 Simulation Seed 1 Alias none Experiment control 1 # and should run the experiment in control.exp once |
| ID:
1 Name: zca #weights variable,connection from id, value): Initial conditions: V -71.81327 hhs 0.98786 nns 0.02457 bbs 0.203517 zzs 0.00141 s_AMPA 0 Associated Variable Values: C_m 1 I_app 0.661914 V_K -90 V_L -70 V_Na 55 g_A 1.4 g_Kdr 6 g_L 0.05 g_Na 35 g_NaP 0.3 g_Z 1 g_syn 0 phi 10 pms 3 pns 4 sigma_a 20 sigma_b -6 sigma_h -7 sigma_k 7 sigma_m 9.5 sigma_n 10 sigma_p 3 sigma_s -2 sigma_z 5 tau_Bs 15 tau_Zs 75 tau_h -40.5 tau_n -27 tau_s 15 theta_a -50 theta_b -80 theta_h -45 theta_m -30 theta_n -35 theta_p -47 theta_s -10 theta_z -39 # Connection Parameters # function values Ainfs_val 0 I_A_val 0 I_Kdr_val 0 I_NaP_val 0 I_Na_val 0 I_appx_val 0 I_z_val 0 Minfs_val 0 Pinfs_val 0 s_inf_val 0 ID: 2 Name: zca #weights variable,connection from id, value): Initial conditions: V -71.81327 hhs 0.98786 nns 0.02457 bbs 0.203517 zzs 0.00141 s_AMPA 0 Associated Variable Values: C_m 1 I_app 0.661914 V_K -90 V_L -70 V_Na 55 g_A 1.4 g_Kdr 6 g_L 0.05 g_Na 35 g_NaP 0.3 g_Z 1 g_syn 0 phi 10 pms 3 pns 4 sigma_a 20 sigma_b -6 sigma_h -7 sigma_k 7 sigma_m 9.5 sigma_n 10 sigma_p 3 sigma_s -2 sigma_z 5 tau_Bs 15 tau_Zs 75 tau_h -40.5 tau_n -27 tau_s 15 theta_a -50 theta_b -80 theta_h -45 theta_m -30 theta_n -35 theta_p -47 theta_s -10 theta_z -39 # Connection Parameters # function values Ainfs_val 0 I_A_val 0 I_Kdr_val 0 I_NaP_val 0 I_Na_val 0 I_appx_val 0 I_z_val 0 Minfs_val 0 Pinfs_val 0 s_inf_val 0 |
| ID:
2 Name: zca #weights variable,connection from id, value): Initial conditions: V -55 hhs 0.98786 nns 0.02457 |
| >
nne_run zca.setup
|
| >
nne_plotPair 1 2
|
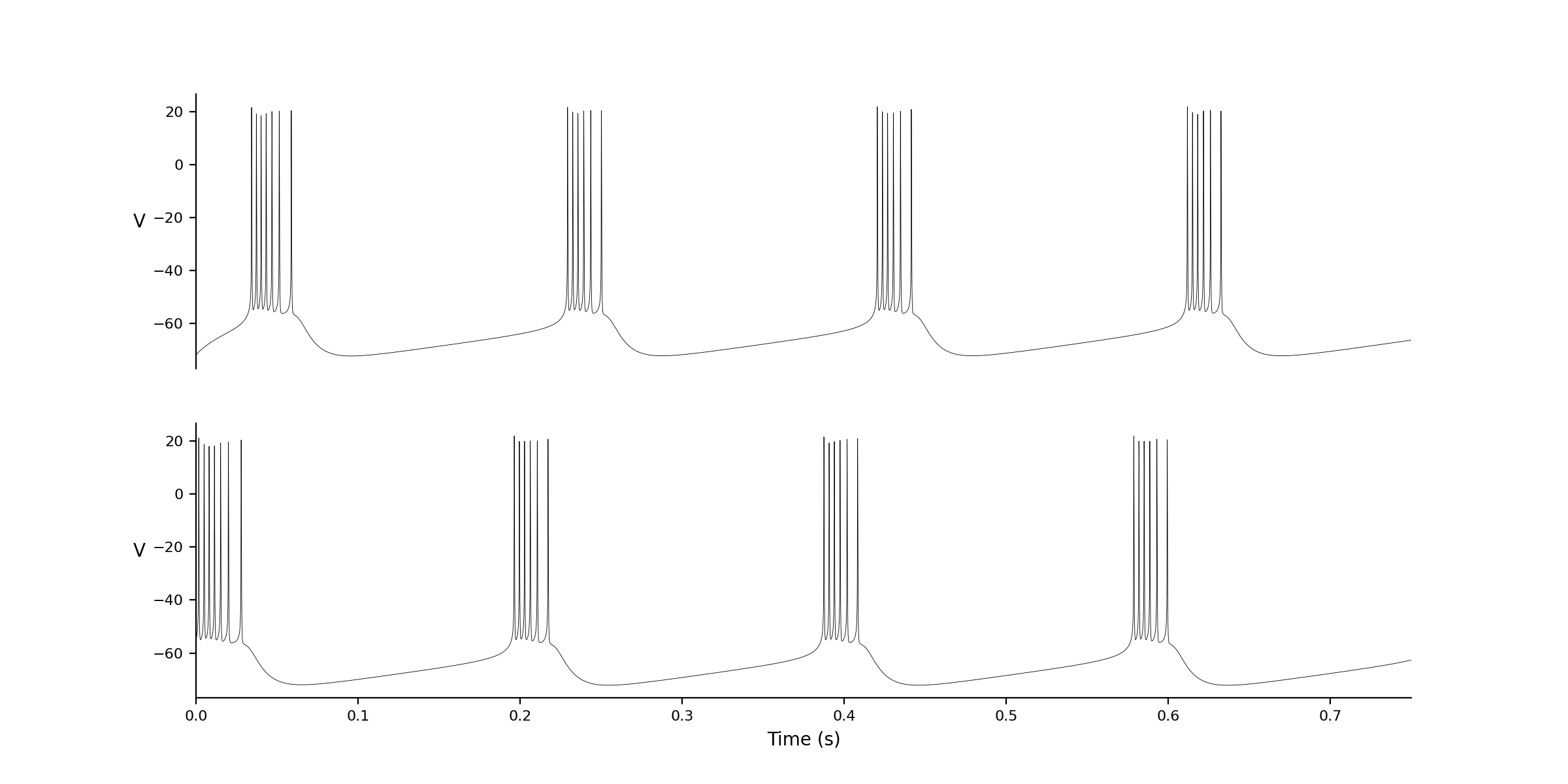
| ID:
1 Name: zca Excitatory Connections To: 2 #weights variable,connection from id, value): Initial conditions: V -71.81327 hhs 0.98786 nns 0.02457 bbs 0.203517 zzs 0.00141 s_AMPA 0 Associated Variable Values: C_m 1 I_app 0.661914 V_K -90 V_L -70 V_Na 55 g_A 1.4 g_Kdr 6 g_L 0.05 g_Na 35 g_NaP 0.3 g_Z 1 g_syn 1 phi 10 pms 3 pns 4 sigma_a 20 sigma_b -6 sigma_h -7 sigma_k 7 sigma_m 9.5 sigma_n 10 sigma_p 3 sigma_s -2 sigma_z 5 tau_Bs 15 tau_Zs 75 tau_h -40.5 tau_n -27 tau_s 15 theta_a -50 theta_b -80 theta_h -45 theta_m -30 theta_n -35 theta_p -47 theta_s -10 theta_z -39 # Connection Parameters # function values Ainfs_val 0 I_A_val 0 I_Kdr_val 0 I_NaP_val 0 I_Na_val 0 I_appx_val 0 I_z_val 0 Minfs_val 0 Pinfs_val 0 s_inf_val 0 ID: 2 Name: zca Excitatory Connections To: 1 #weights variable,connection from id, value): Initial conditions: V -55 hhs 0.98786 nns 0.02457 bbs 0.203517 zzs 0.00141 s_AMPA 0 Associated Variable Values: C_m 1 I_app 0.661914 V_K -90 V_L -70 V_Na 55 g_A 1.4 g_Kdr 6 g_L 0.05 g_Na 35 g_NaP 0.3 g_Z 1 g_syn 1 phi 10 pms 3 pns 4 sigma_a 20 sigma_b -6 sigma_h -7 sigma_k 7 sigma_m 9.5 sigma_n 10 sigma_p 3 sigma_s -2 sigma_z 5 tau_Bs 15 tau_Zs 75 tau_h -40.5 tau_n -27 tau_s 15 theta_a -50 theta_b -80 theta_h -45 theta_m -30 theta_n -35 theta_p -47 theta_s -10 theta_z -39 # Connection Parameters # function values Ainfs_val 0 I_A_val 0 I_Kdr_val 0 I_NaP_val 0 I_Na_val 0 I_appx_val 0 I_z_val 0 Minfs_val 0 Pinfs_val 0 s_inf_val 0 |
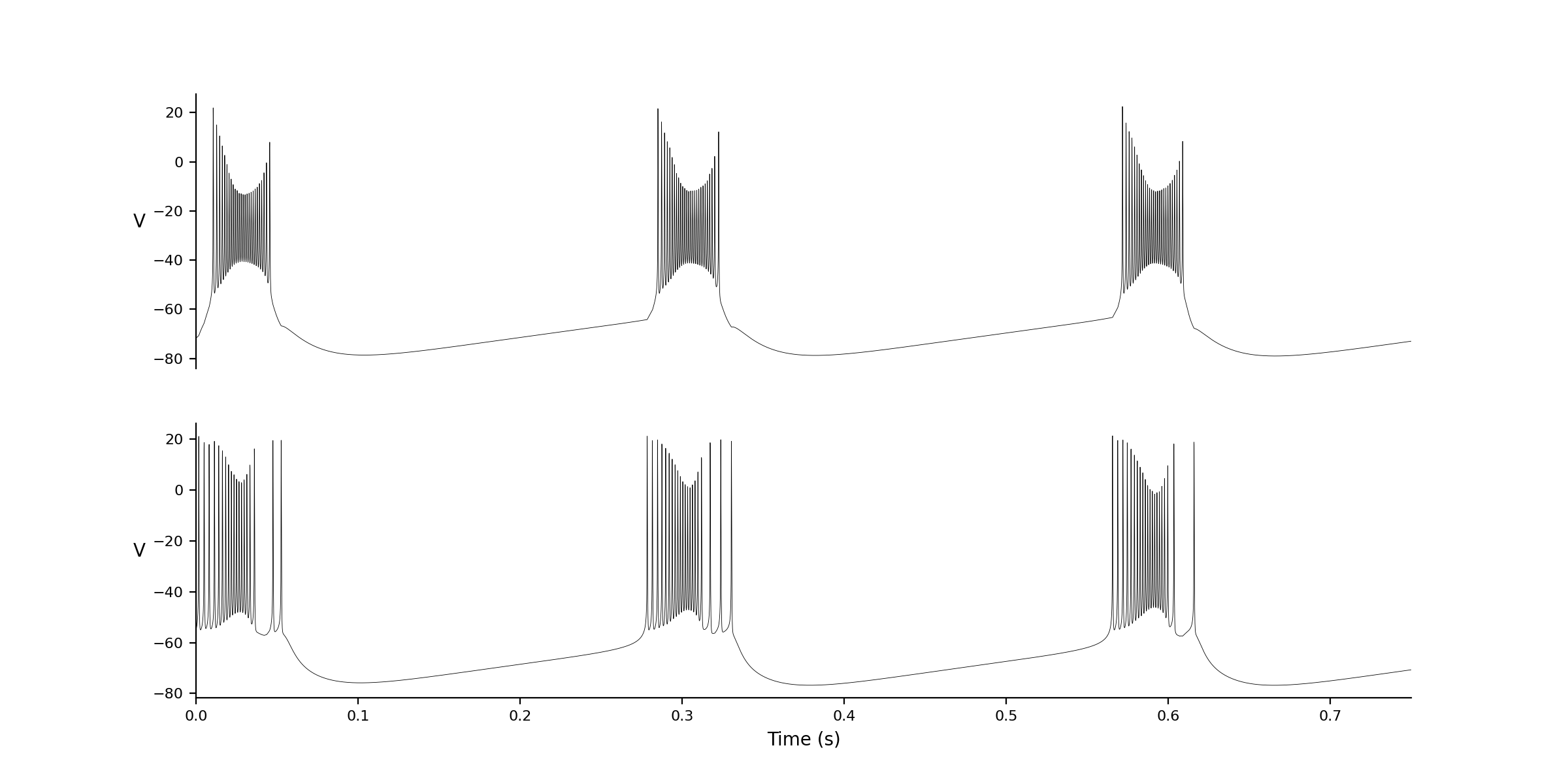
| #
string user uses to refer to this connection type :
list of variables that are connected by this connection
type, e.g., Excitatory: s_AMPA s_NMDA Inhibitory: s_GLY s_GABA Electrical: V |